PtrHash
Minimal Perfect Hash Functions Link to heading
A set of \(n\) keys Link to heading
A hash function Link to heading
A hash function: collisions! Link to heading
A hash function: injective / perfect Link to heading
A hash function: bijective / minimal & perfect Link to heading
Problem statement Link to heading
Given a set of \(n\) keys \(K\subseteq \mathbb K\), build a function \(h\) satisfying
- \(h(K) = \{0, \dots, n-1\}\): \(h\) is minimal & perfect.
Why? Array of \(n\) values as space-efficient value store.
Solutions take \(\log_2(e) {=} 1.443\) to \(3\) bits/key.
Goals of PtrHash:
- at most 3 bits/key (you probably store ≥16 bit values anyway),
- fast to evaluate,
- fast to construct.
Previous & parallel work Link to heading
- FCH: Fox, Chen, and Heath (1992)
- CHD: Belazzougui, Botelho, and Dietzfelbinger (2009)
- PTHash: Pibiri and Trani (2021)
- PHOBIC: Hermann et al. (2024)
PHast: Beling and Sanders (2025)
Consensus: Lehmann, Sanders, et al. (2025)
Survey: Lehmann, Mueller, et al. (2025)
\[ \newcommand{\lo}{\mathsf{lo}} \newcommand{\hi}{\mathsf{hi}} \]
Naive: try seeded hashes Link to heading
Naive: try seeded hashes Link to heading
Naive: try seeded hashes Link to heading
Naive: \(n^n / n!\approx e^n\) tries, \(n\cdot\log_2 e\) bits Link to heading
Construction Link to heading
Build \(h\) incrementally using buckets Link to heading
Build \(h\) incrementally Link to heading
Build \(h\) incrementally Link to heading
Build \(h\) incrementally Link to heading
Build \(h\) incrementally Link to heading
Build \(h\) incrementally Link to heading
Build \(h\) incrementally Link to heading
Build \(h\) incrementally Link to heading
Build \(h\) incrementally Link to heading
Build \(h\) incrementally Link to heading
Build \(h\) incrementally Link to heading
Build \(h\) incrementally Link to heading
Build \(h\) incrementally Link to heading
Better: assign buckets from large to small Link to heading
Skew bucket sizes Link to heading
Remapping: avoiding the hard part Link to heading
Remapping: add \(1\%\) extra slots Link to heading
Remapping: add \(1\%\) extra slots Link to heading
Remapping: map extra slots back Link to heading
Remapping: map extra slots back Link to heading
Queries Link to heading
Query: 1. Hash key Link to heading
Query: 2. Lookup bucket Link to heading
Query: 3. Compute slot Link to heading
PTHash Link to heading
- Iterate seed values (pilots) until a hit is found.
- Dictionary compression to handle large values.
PtrHash Link to heading
- Iterate seed values (pilots) until a hit is found.
- Dictionary compression to handle large values.
- “Inlined” 8-bit pilots for fast lookup
- Handle impossible buckets by evicting others
- When none of the \(2^8=256\) seeds are collision free, choose the one with minimal number of collisions, and break ties towards evicting small buckets.
- Eviction: “unassign” a seed, and put the bucket back in the queue.
- Distributes the entropy from the hard buckets over all seeds.
<no animation here :(>
\(\varepsilon\) cost sharding Link to heading
- \(S\) shards
- PTHash, PHOBIC: shards of expected size \(a = n/S\), real size \(a_i\)
- \(a / \lambda\) buckets per shard, \(a_i/\alpha\) slots per shard
- Store shard offsets
- PtrHash: \(a / \lambda\) buckets per shard, \(a /\alpha\) slots per shard
- Direct indexing
- Shard size follows from Sebastiano’s formula
PtrHash with array vs VFUNC Link to heading
- VFUNC, storing \(b\) bit values:
- 10% memory overhead: \(b n + 0.1 b n\)
- 3 independent/parallel reads
- PtrHash:
- 3 bits overhead: \(b n + 3n\)
- 2 dependent/sequential reads
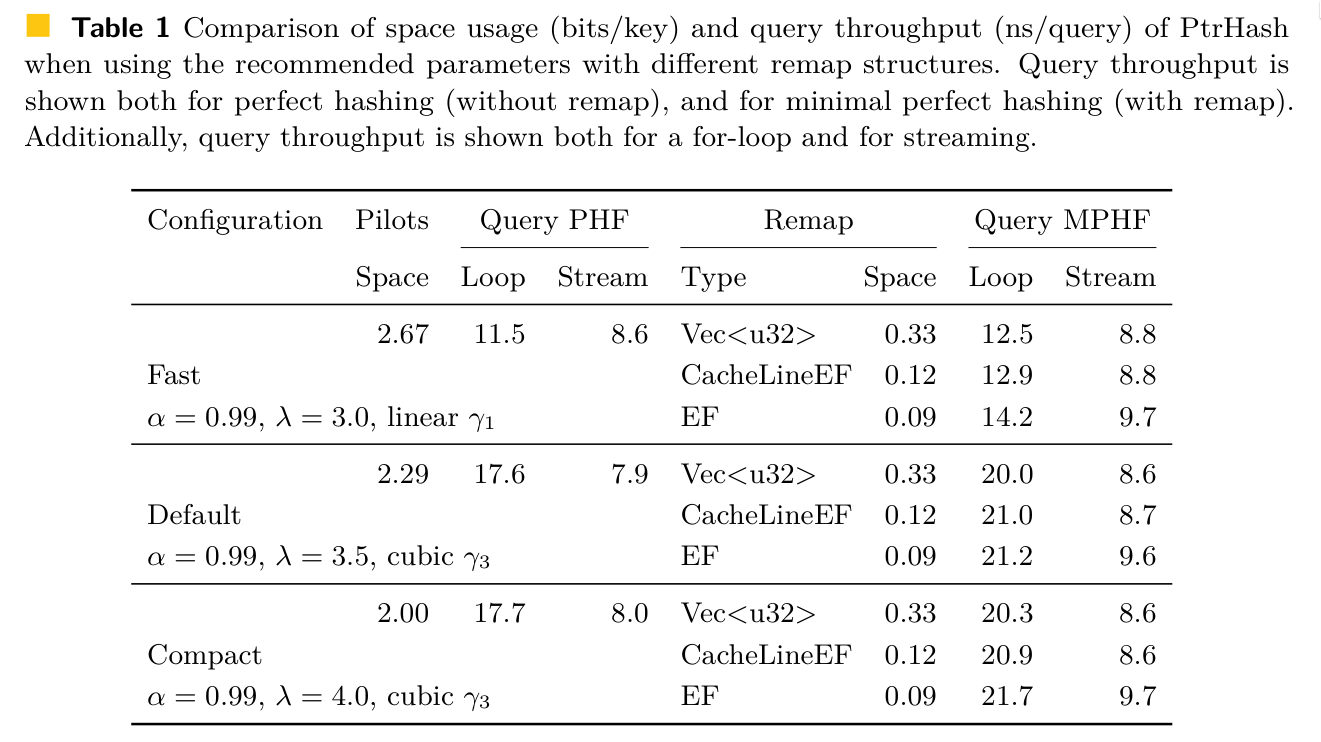
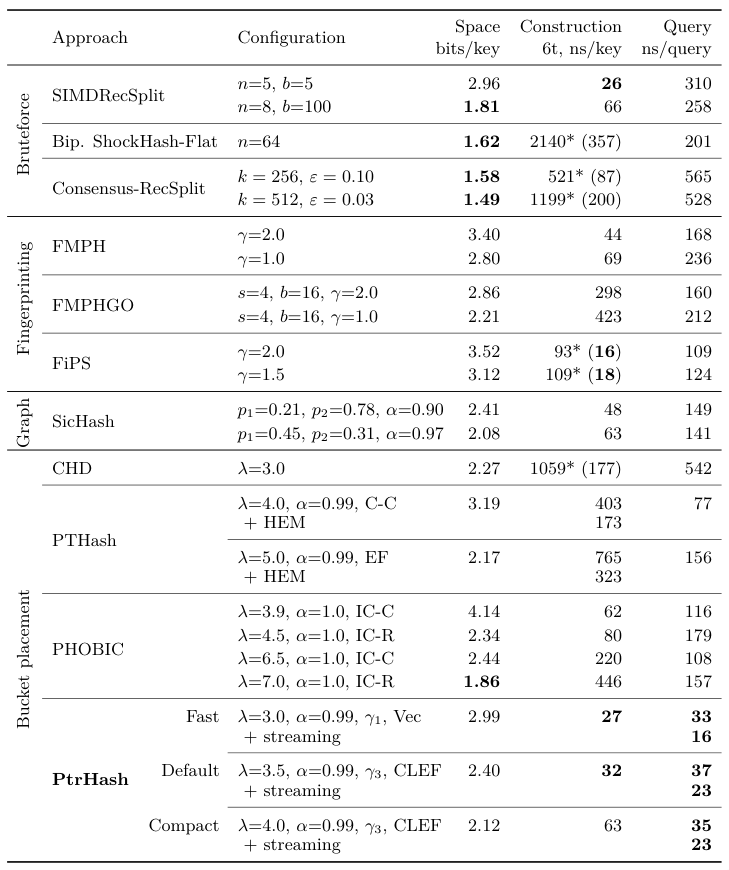
Results Link to heading
Construction time vs. size Link to heading
Comparison Link to heading
- 300M variable length string keys
- <3 bits/key
- fastest queries by 2x to 4x
- also fast construction
Queries: looping, batching, streaming Link to heading
A simple loop Link to heading
- Setup: PtrHash on \(10^9\) integer keys; 300 MB.
- Fact: Most queries need uncached data from RAM.
- Fact: Reading RAM takes 80 ns
- Question: how long does this take? (>90, ≈80, 40-60, <30)
| |
- Answer: 12 ns!
- The CPU works on 7 iterations in parallel!
- To measure latency:
| |
Batching & Streaming Link to heading
Batching:
| |
Streaming:
| |
Batching with Prefetching Link to heading
Batching with Prefetching Link to heading
Batching & Prefetching Link to heading
Multithreading Link to heading
Conclusions Link to heading
- Simple \(\implies\) fast
- Partition data into cache-sized chunks
- Streaming & prefetching for max throughput
Brought to you by Link to heading

Bonus slides Link to heading
Simple operations Link to heading
- Hashing via \[h(x) = \lo(C\cdot x) = (C\cdot x)\mod 2^{64}.\]
- Quadratic bucket function \[\gamma(x) = \hi(x\cdot x) = \lfloor x^2/2^{64}\rfloor.\]
- Use fastmod for \(\mod n\)
Overview Link to heading
Parameters Link to heading