Optimal Throughput Bioinformatics
PhD Defense
Ragnar {Groot Koerkamp}
April 10, 2025
What is bioinformatics?
For today: DNA
Zephyris, CC BY-SA 3.0 Wikipedia
Also DNA

Human chromosomes
DNA, according to me
A C G T
00 01 10 11
Covid

A virus: SARS-CoV-2, aka COVID-19
Why bioinformatics?
1970-2000: Floppy disks - MB - Bacteria

Floppy
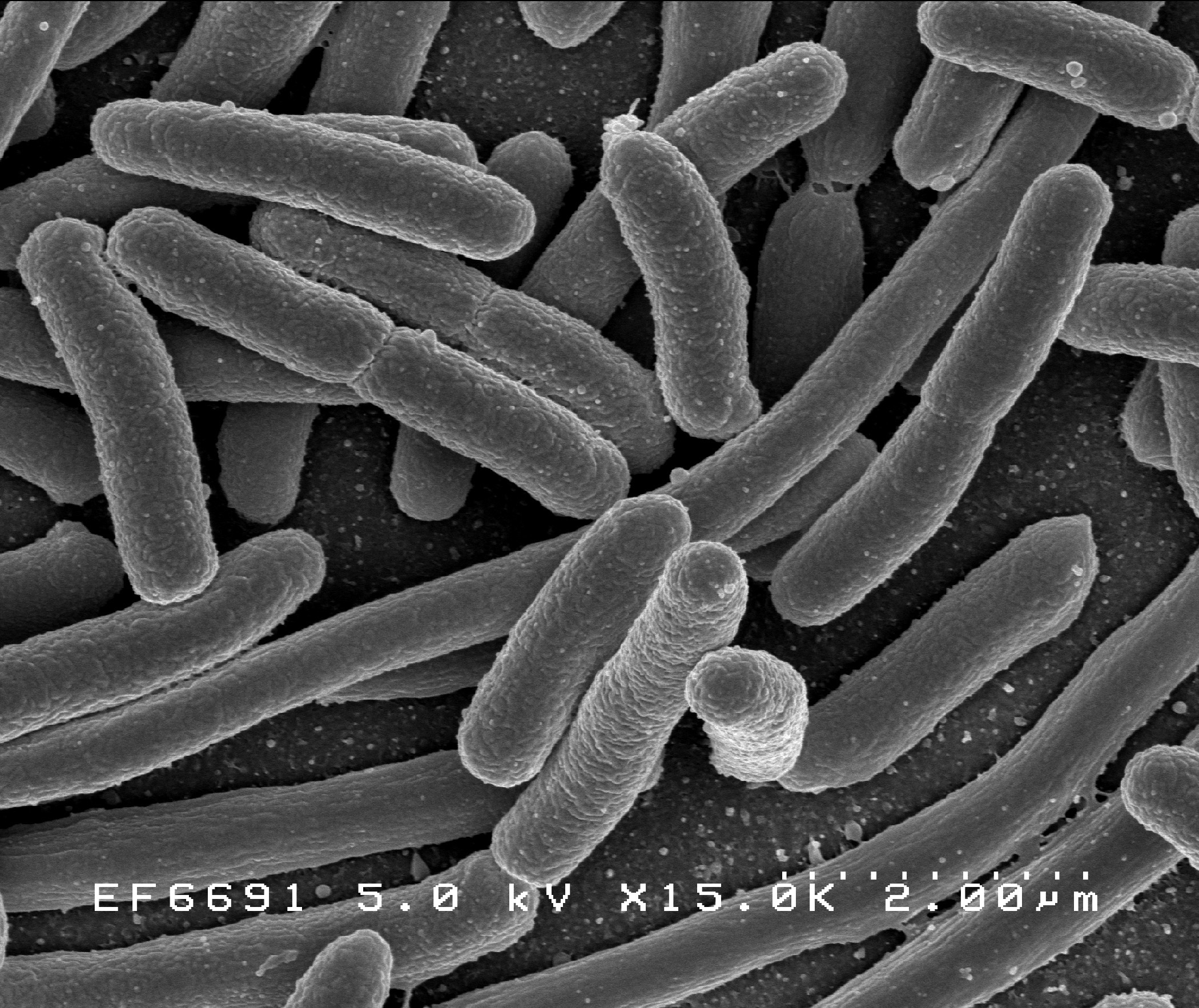
E. coli bacteria (1997)
2000-2020: USB stick - GB - Human Genome

USB stick

Human genome (2001)
2010-2020: Hard Drive - TB - RefSeq

Hard drive

RefSeq
2025: Data Center - PB - SRA

Data Center
PhonlamaiPhoto | istockphoto.com
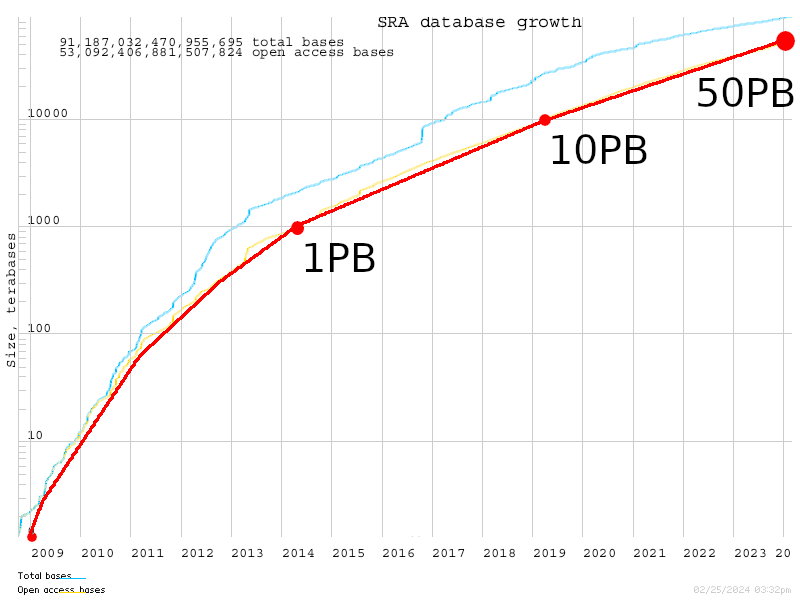
Growth of SRA
Goal:
Fast code
Goal:
High throughput code
Goal:
Optimal throughput code
What is high troughput code?
- Complexity
Few operations:
\(\quad O(n^2)\quad\longleftrightarrow\quad O(n)\)
- Efficiency
Fast operations:
memory read, 100 ns \(\quad\longleftrightarrow\quad\) 0.1 ns, addition
- Implementation
Parallel operations:
SIMD, instruction-level parallelism
Papers
Part 1: Pairwise Alignment
Part 2: Minimizers
Part 3: Optimal Throughput
Additional
- [1] A*PA: Exact Global Alignment Using A* with Chaining Seed Heuristic and Match Pruning.
RGK and Pesho Ivanov, Bioinformatics 2024. - [2] A*PA2: Up to 19x Faster Exact Global Alignment.
RGK. WABI 2024.
- [3] The Mod-Minimizer: A Simple and Efficient Sampling Algorithm for Long k-mers.
RGK and Giulio Ermanno Pibiri. WABI 2024. - [4] Forward Sampling Scheme Density Lower Bound.
Bryce Kille, RGK, et al. Bioinformatics 2024. - [5] The Open-Closed Mod-Minimizer Algorithm.
RGK, Daniel Liu, and Giulio Ermanno Pibiri. AMB 2025.
- [6] SimdMinimizers: Computing Random Minimizers, Fast.
RGK and Igor Martayan. SEA 2025. - [7] PtrHash: Minimal Perfect Hashing at RAM Throughput.
RGK. SEA 2025.
- [8] U-index: A Universal Indexing Framework for Matching Long Patterns.
Lorraine Ayad, Gabriele Fici, RGK, Rob Patro, Giulio Ermanno Pibiri, and Solon Pissis. SEA 2025.
Problem 1: Pairwise Alignment
Covid – \(\alpha\), December 2020

Sars-CoV-2, alpha variant, December 2020
Covid – \(\omicron\), December 2021

Sars-CoV-2, omicron variant, December 2021
Pairwise alignment
- Find the mutations between two sequences
Dynamic programming
Dynamic programming
Dynamic programming
Dynamic programming
Dynamic programming
Needleman-Wunsch – Quadratic \(O(n^2)\)

Dijkstra – \(O(ns)\)

Diagonal transition – \(O(n + s^2)\)

A*PA\({}^{1}\) – near-linear, \(3\times\) faster on similar seqs

[1] Exact Global Alignment Using A* with Chaining Seed Heuristic and Match Pruning.
RGK and Pesho Ivanov, Bioinformatics 2024.
A*PA\({}^{1}\) – not quite linear

[1] Exact Global Alignment Using A* with Chaining Seed Heuristic and Match Pruning.
RGK and Pesho Ivanov, Bioinformatics 2024.
A*PA: Great complexity – terrible efficiency
Band Doubling – \(O(ns)\)

A*PA2\({}^{2}\) – good efficiency: up to \(19\times\) faster

[2] A*PA2: Up to 19x Faster Exact Global Alignment.
RGK. WABI 2024.
Problem 2: Minimizers – lossy compression
Lossy compression
original
Lossy compression
50%
Lossy compression
25%
Lossy compression

12%
Lossy compression

6%
Sampling k-mers

\(k=10\), \(w=1000\)
Minimizer definition
\(k\)-mer size: \(k=3\)
Window guarantee: \(w=4\)
Length \(w+k-1=6\) window of \(w\) \(k\)-mers
Minimizer scheme:
\[
f: \Sigma^{w+k-1} \mapsto \{0, 1, 2, \dots, w-1\}.
\]
Used for compression and hashing.
Minimizer example
Minimizer example
Minimizer example
Minimizer example
Minimizer example
Minimizer example
Minimizer density
Density: expected fraction of sampled \(k\)-mers.
Here: \(3/9=0.33\)
Sampling k-mers

\(k=10\), \(w=500\)
Sampling k-mers

\(k=10\), \(w=250\)
Sampling k-mers

\(k=10\), \(w=100\)
Sampling k-mers

\(k=10\), \(w=50\)
Sampling k-mers

\(k=10\), \(w=25\)
Sampling k-mers

\(k=10\), \(w=100\)
Sampling k-mers

\(k=11\), \(w=100\)
Sampling k-mers

\(k=12\), \(w=100\)
Sampling k-mers

\(k=20\), \(w=100\)
Sampling k-mers

\(k=30\), \(w=100\)
Goal: Minimize the number of sampled \(k\)-mers
Density plots – before (\(w=24\))
The Mod-Minimizer\({}^{3}\)
[3] The Mod-Minimizer: A Simple and Efficient Sampling Algorithm for Long k-mers. RGK and Giulio Ermanno Pibiri. WABI 2024.
A Near-Tight Lower Bound\({}^{4}\)
[4] Forward sampling scheme density lower bound. Bryce Kille, RGK, et al. Bioinformatics 2024.
Near-optimal schemes for small \(k\)
(Unpublished)
Extended mod-minimizer\({}^{5}\)
[5] The Open-closed mod-minimizer Algorithm. RGK, Daniel Liu, and Giulio Ermanno Pibiri. AMB 2025.
Proving The Lower Bound
Suppose \(k=1\), and consider a cycle of \(w+1\) characters.
Eg for \(w=4\):ABCDEABCDEABCDE
Suppose we only sample 1 character:
ABCDEABCDEABCDE
The distance between samples is \(5 > w\)!
- Thus, we need at least two samples.
- Conclusion: the density is at least \(2/(w+1)\).
Conclusion: high troughput code matters!
- Complexity
Few operations:
A*PA: \(\quad O(n^2)\quad \mapsto\quad \ ``O(n)\text{''}\)
Provably near-optimal minimizer schemes.
- Efficiency
Fast operations:
A*PA2 is up to 500x more efficient than A*PA; up to 19x faster than other methods.
- Implementation
Parallel operations:
A*PA2 uses SIMD and instruction-level parallelism.
Outlook
- Optimize all the code.
- Minimizers are not yet a fully solved problem.
- Proving optimality is hard.
Thanks!
Propositions
- Complexity theory’s days are numbered.
- \(\log \log n \leq 6\)
- Succinct data structures are overrated.
- There is beauty in mathematical perfection.
- Too many PhDs are wasted shaving of small factors of complexities that will never be practical.
- If a paper starts with “faster methods are needed”, it must talk about the implementation.
- Fast code must exploit assumptions on the input.
- Fast code puts requirements on the input format.
- Optimizing ugly code is a waste of time.
- Assembly is not scary.
Extra: Pairwise alignment
A*PA: comparison
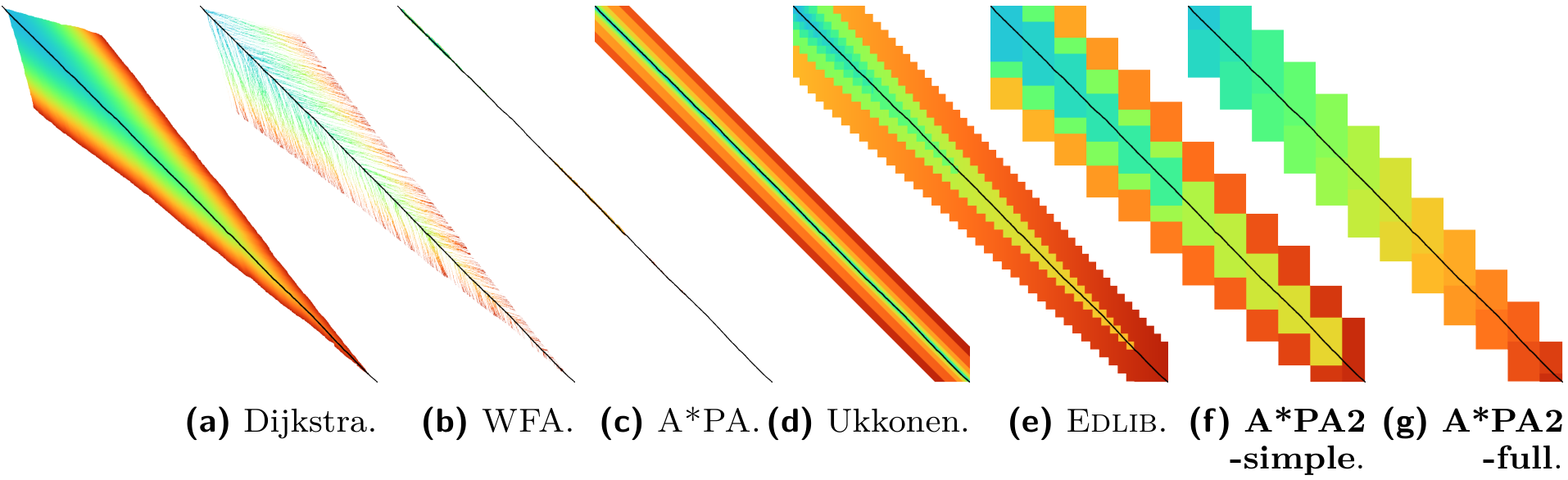
A*PA heuristics
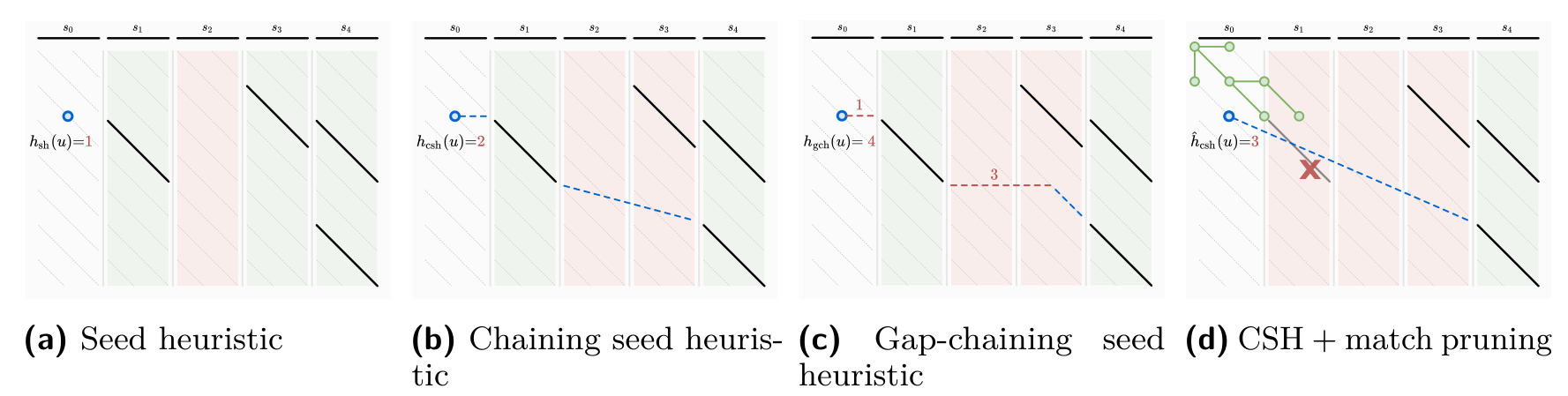
A*PA: seed heuristic
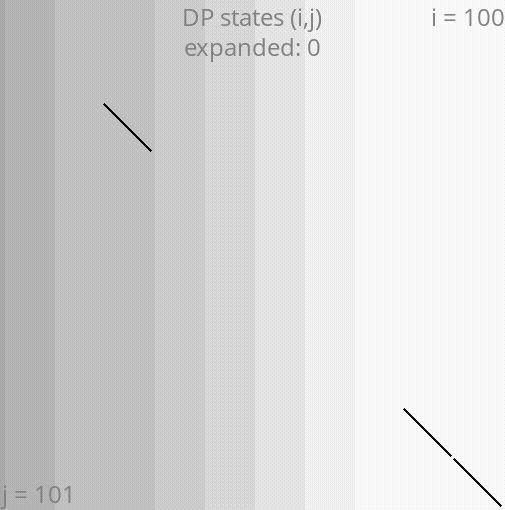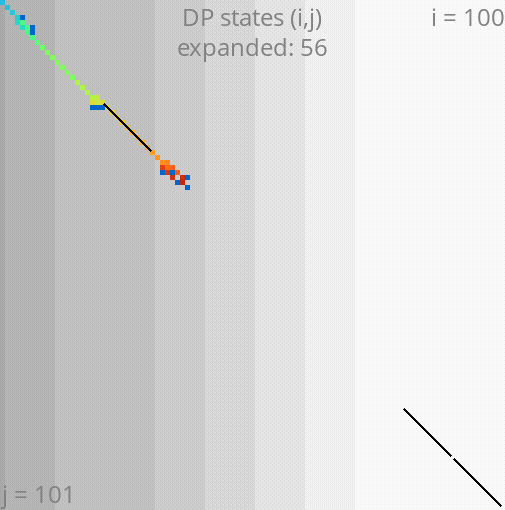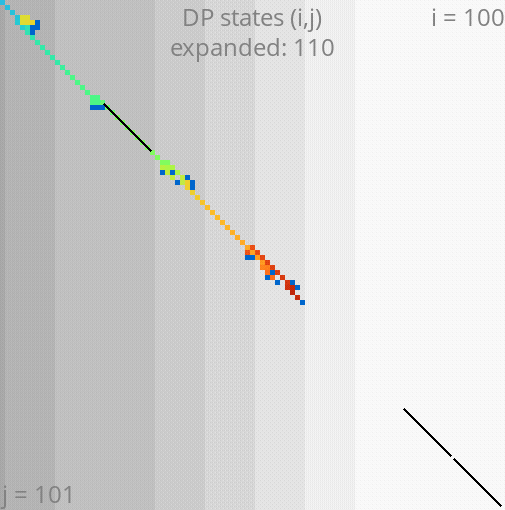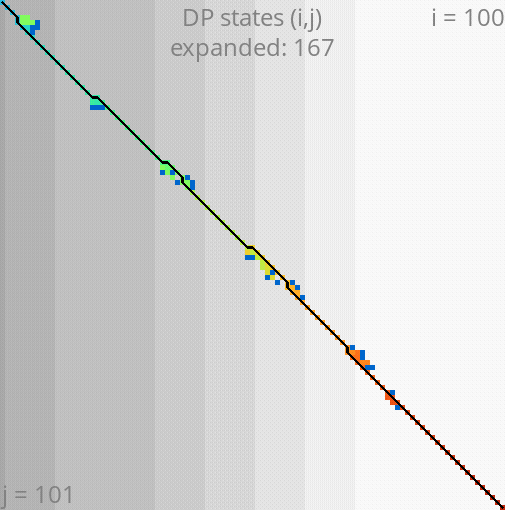
A*PA: gap-chaining seed heuristic
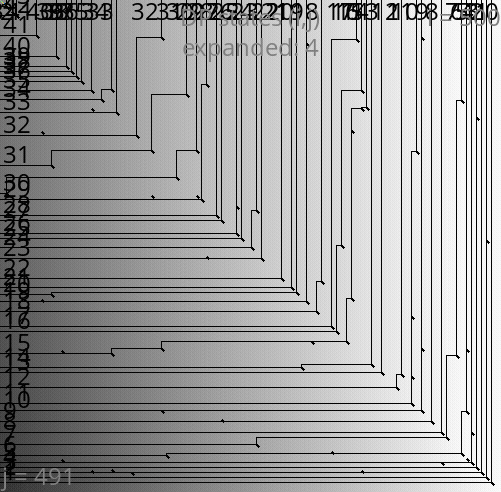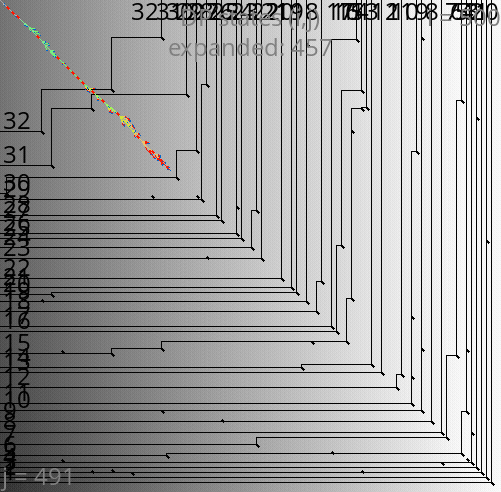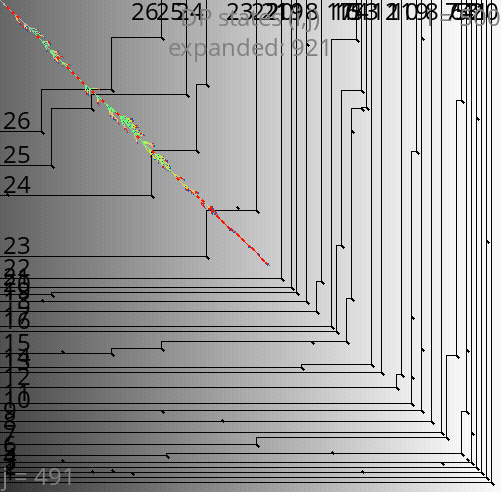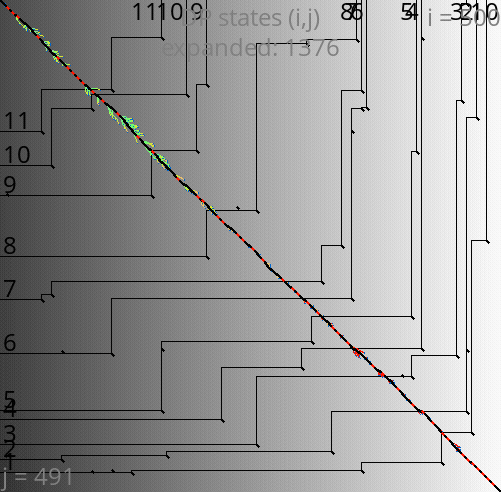
A*PA: contours
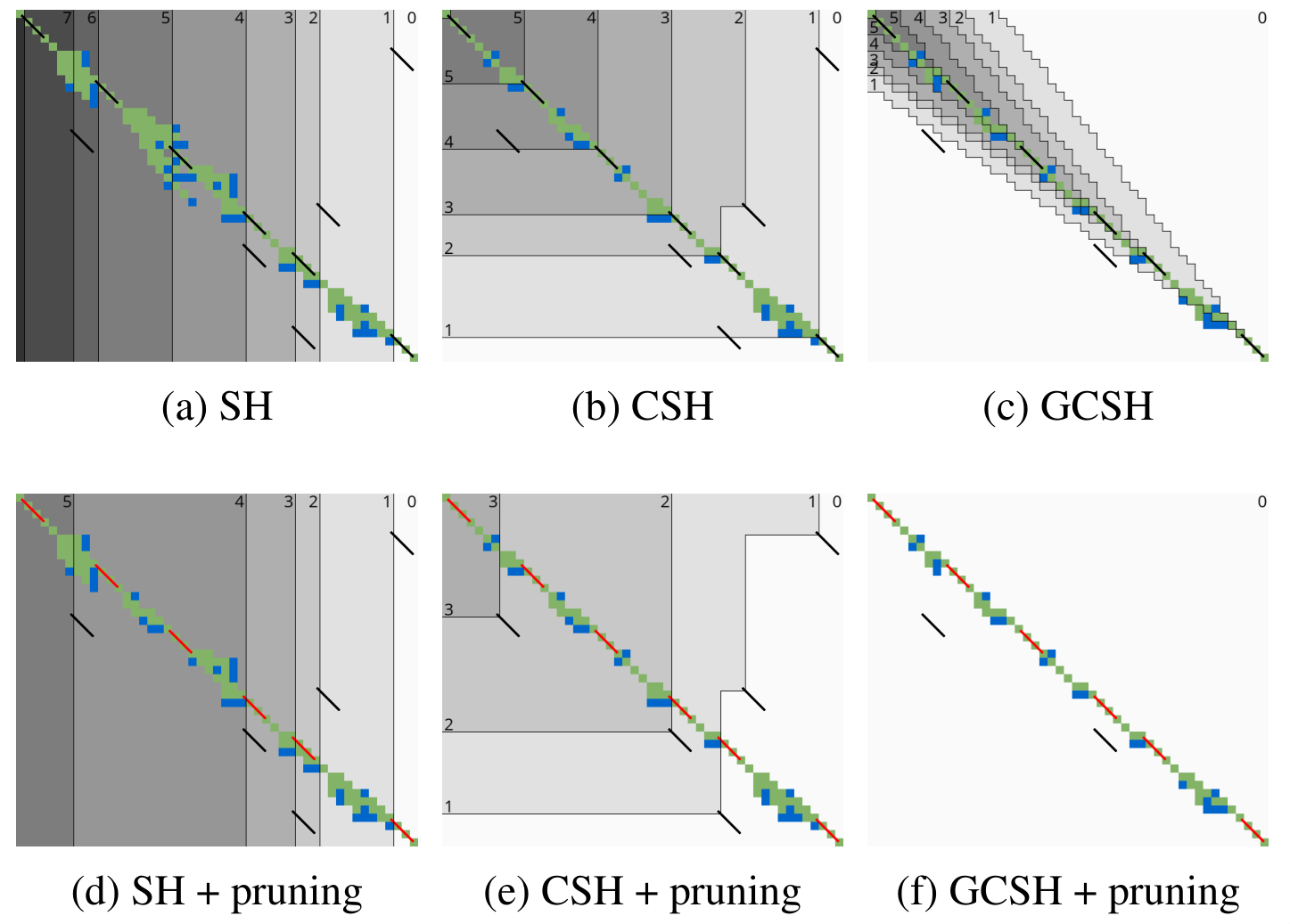
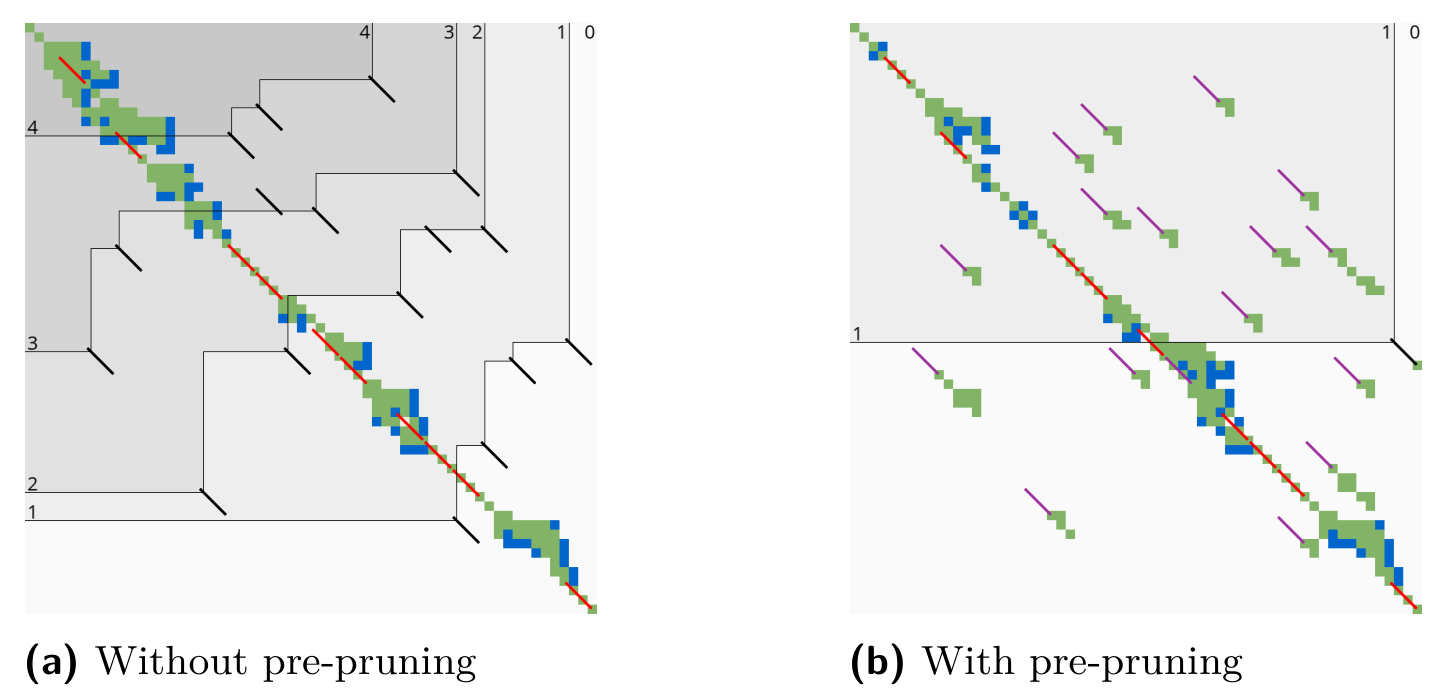
A*PA2: pre-pruning
A*PA2: results (real data)
A*PA2: results (synthetic)
Alignment modes
Cost models
Semi-global variants
Text searching
Skip-cost
Skip-cost
Block-computation results
Semi-global A*PA

Seed-chain-extend
A*PA: table
Extra: Minimizers
Minimizer schemes
- Lexicographic: choose lexicographic smallest \(k\)-mer
- Random mini: choose \(k\)-mer with smallest hash
- ABB: choose A, followed by most non-A
- ABB+: break ties via random hash
- Sus-anchor: choose the position of the smallest unique suffix
- “smallest”: where the first character is inverted.
Large alphabet
Lower bound
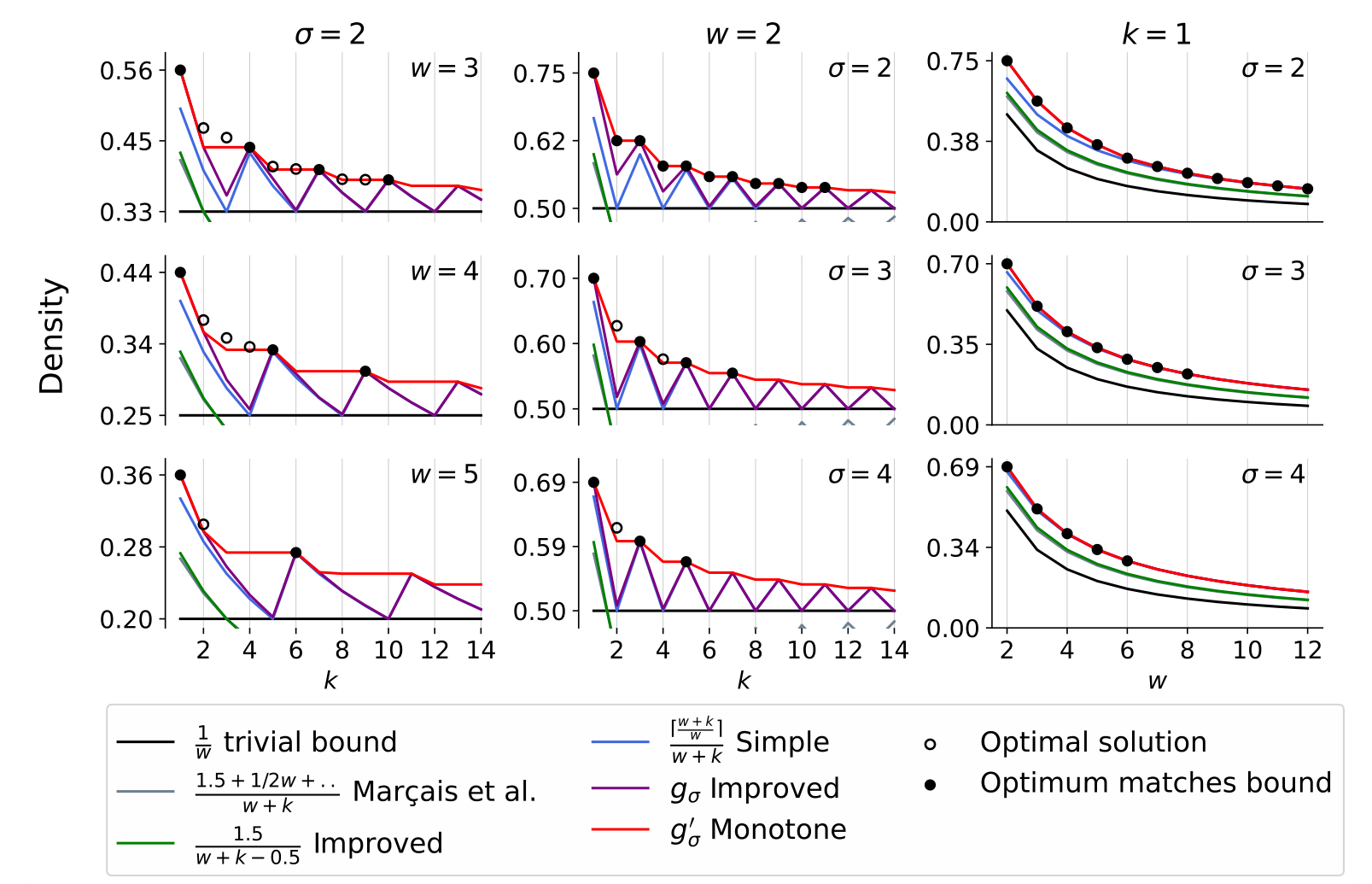
Mod-minimizer
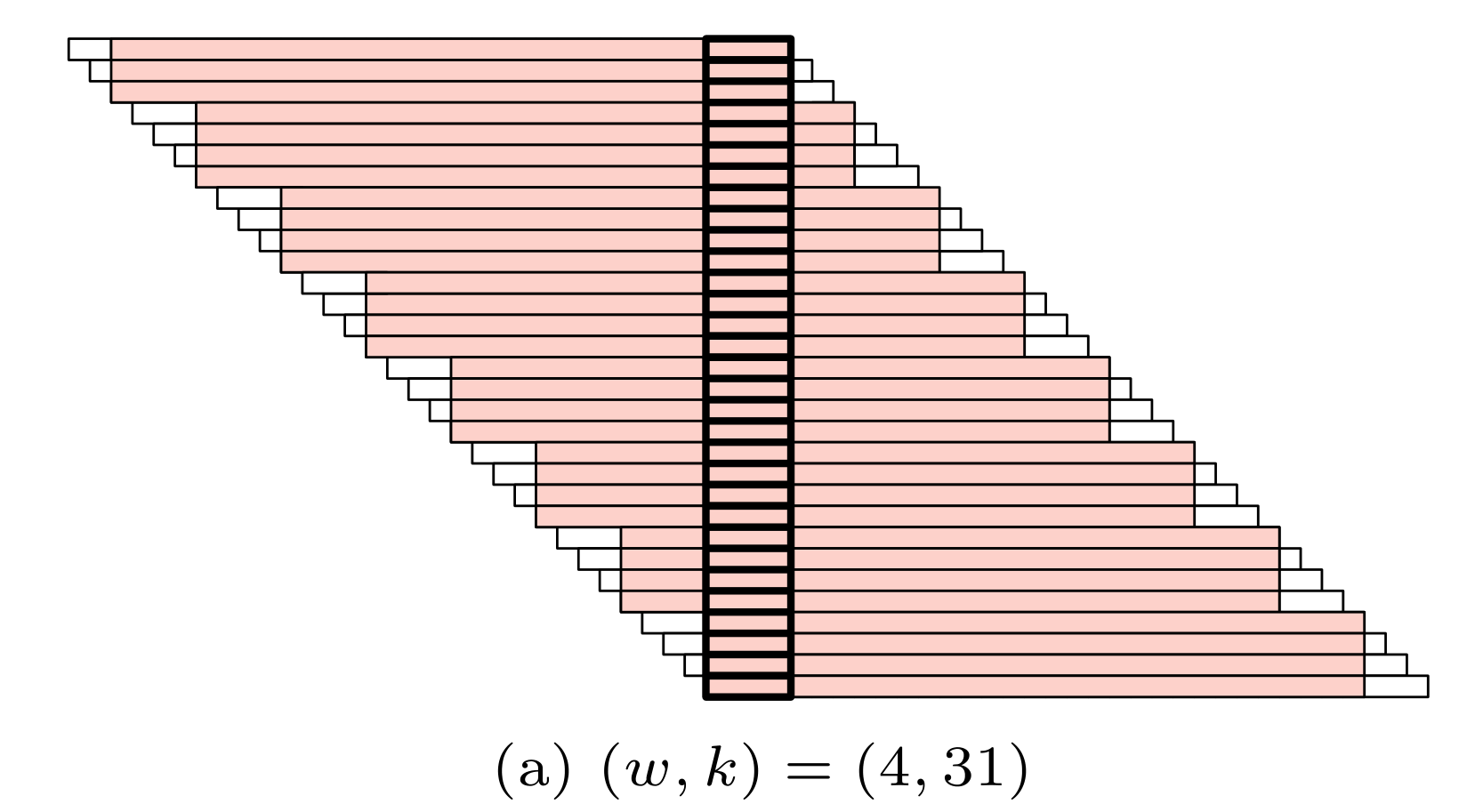
Mod-minimizer
Mod-minimizer
Mod-minimizer
Mod-minimizer
Mod-minimizer
Selection schemes
Extra: PtrHash
PtrHash: overview
PtrHash: Cacheline Elias Fano
PtrHash: Bucket Functions
PtrHash: Construction
PtrHash: Results
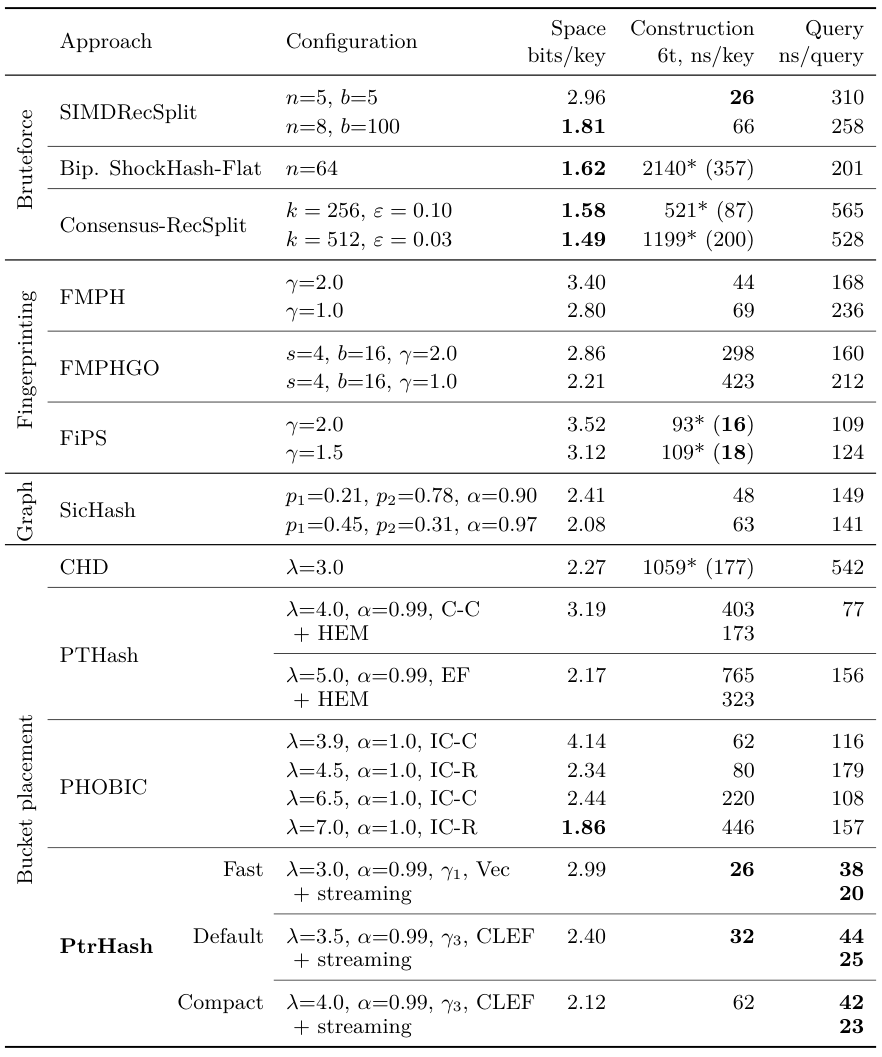
PtrHash: Optimal Throughput